Nanobody screening
Nanobody Screening Based on Yeast Surface Display (Q&A)
What is a nanobody and what are its uses?
Nanobody is a single-domain antibody composed solely of the variable region of the heavy chain, also known as VHH. With a molecular weight of 13-14 kD, it is the smallest naturally occurring antigen-binding fragment.
Compared with traditional antibodies, nanobodies have characteristics such as higher affinity and stability, ease of preparation, low immunogenicity, good solubility, and strong penetration. They have broad applications in cancers, autoimmune diseases, respiratory diseases, and hematological diseases.
How does yeast surface display work?
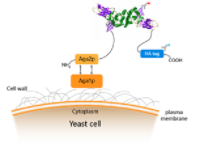
The yeast surface display uses the α-agglutinin system. The α-agglutinin is composed of two core subunits, Aga1 and Aga2. In this system, the gene encoding the Aga1 protein is stably integrated into the yeast chromosome, while Aga2 is cloned into a circular yeast display plasmid vector. After induced expression by galactose, Aga1 is anchored to the cell wall, and the foreign protein is expressed in fusion with Aga2. With the help of signal peptide, Aga2 is expressed and secreted, where Aga2 covalently links to Aga1 through two disulfide bonds to form a covalent complex. In this way, the cell surface display of the target protein is achieved.
Which library is used for screening?
We use a fully synthetic nanobody library. We use a fully synthetic nanobody library. The yeast strain is Saccharomyces cerevisiae EBY100. The display plasmid is pYDS649. The library size is approximately 5.6×10⁸.
What is the screening strategy?
We use a combination of magnetic-activated cell sorting (MACS) and fluorescence- activated cell sorting (FACS) to screen for nanobodies.
What is the affinity range between the resulting nanobodies and antigens?
In general, the affinity between nanobodies and antigens can reach the nanomolar (nM) level.
How much antigen protein is required for screening?
Taking a 50 kD protein as an example, approximately 0.5 mg of protein is required for MACS and FACS. The amount of antigen protein needed for size-exclusion chromatography (SEC) and SPR verification is correlated to the number of nanobodies to be verified. Approximately 0.1 mg of antigen protein is required for the verification of each nanobody by SEC and SPR.
What kind of labeling is required for the antigen proteins?
For antigen proteins used in MACS and FACS, options include FITC labeling, biotinylation, or adding a Flag tag, etc.
Biotinylated or unlabeled antigen proteins can be used for SEC and SPR verification.
How long does the screening cycle take?
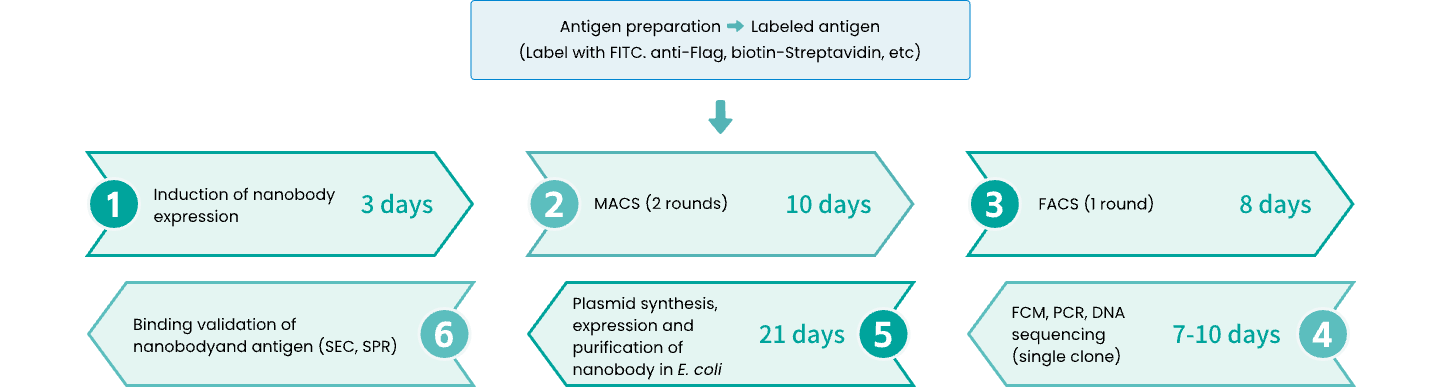
Generally, it takes 2.5 to 3 months (including MACS, FACS, nanobody plasmid synthesis, protein purification, binding verification, etc.).
Schematic diagram of the screening process
Screening instruments
-
MACS


-
Flow-Cytometry


Examples
Results of flow-cytometry cell sorting

SEC and SPR verification

Nanobody-assisted structure determination by Cryo-EM

Nanobody for membrane protein



