Characterization Of Drug Delivery Systems (LNPs, AAVs, Exosomes, etc.)
Advantages
-
Low sample volume requirement (30 μL,1 mg/mL concentration)
-
No sample pre-treatment required
-
Fast Turnaround (results available as soon as the same day)
-
Real morphological information obtained (especially regarding internal structure)
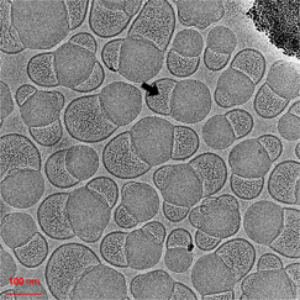
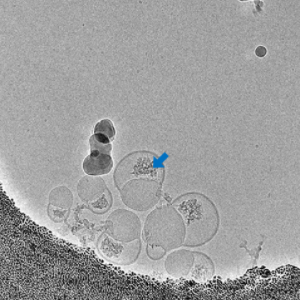
Morphology
-
API-liposome

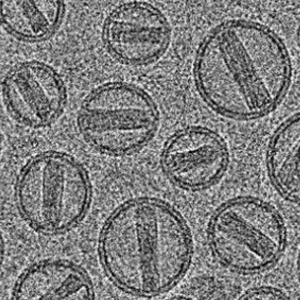
-
mRNA-LNP

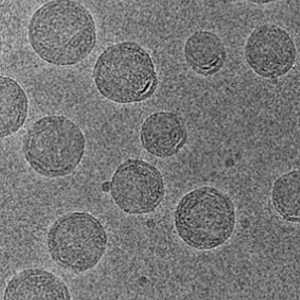
-
Exosome

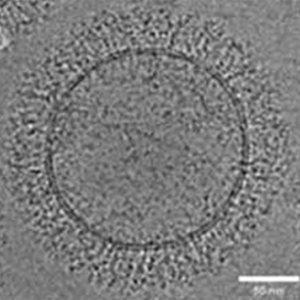
-
AAV

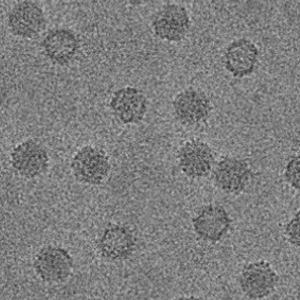
Cryo-EM Morphological Characterization of Drug Delivery Systems
Particle Extraction and Size Distribution Statistics Based on Deep Learning
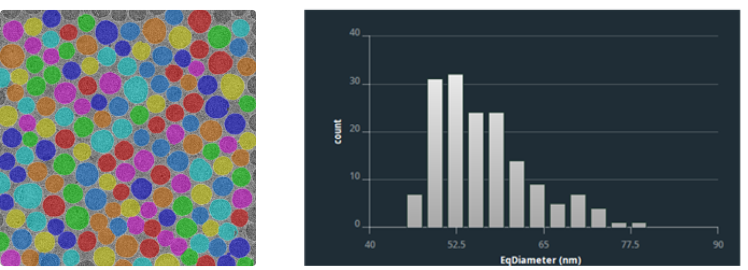
Size Distribution Statistics of Drug Delivery Systems
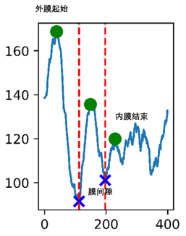
Membrane Thickness Analysis
Membrane Thickness Analysis of Drug Delivery Systems
Empty-to-Full Ratio Statistics
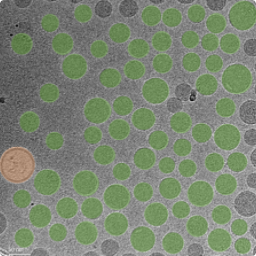
Empty-to-Full Ratio for Lipid-based Samples
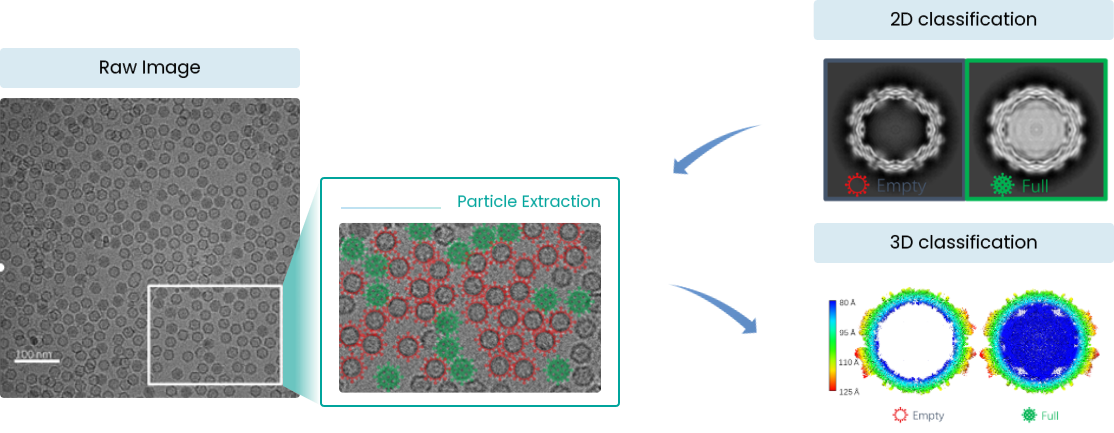
Empty-to-Full Ratio for AAV Samples
mRNA Localization by Thionine Staining
-
Before Staining

![]()
-
After Staining

![]()
-
Before Staining

![]()
-
After Staining

![]()
Localization of mRNA in LNP Samples by Thionine Staining
3D Reconstruction by Cryo Electron Tomography (cryo-ET)

Reconstruction Results of Multivesicular Liposomes by cryo-ET
Q&A
What is the typical turnaround time for sample analysis?
The sample preparation will be conducted on the day when sample is received, and observation results can be obtained on the same day when the sample is scheduled for the cryo-electron microscope. Statistical analysis results will require an additional 2-3 days. If the machine time is reserved in advance before submitting the sample, the waiting time can be reduced.
What are the requirements for the LNP samples?
The total lipid concentration should generally be greater than 1 mg/mL, and the sucrose content should be as low as possible, typically not exceeding 10%.
Can freeze-dried powder samples be observed?
Yes,but freeze-dried powder samples often have high sucrose content or low concentration after reconstitution. Typically, the samples need to be dialyzed or concentrated after reconstitution.
What are the advantages of using cryo-electron microscopy to characterize drug delivery carriers?
Cryo-EM provides direct imaging information of the sample, especially internal structural details. The cryo-preparation preserves the native morphology of the sample, allowing for accurate particle size distribution.
Are the results from DLS and cryo-EM consistent?
Since DLS measures the hydrodynamic diameter in liquid, while cryo-EM provides the actual particle size, the DLS results will be slightly larger. However,the overall distribution trends are consistent.
I heard that cryo-EM can achieve a resolution of 3Å.Can mRNA molecules inside LNP samples be directly observed in EM images?
The resolution we refer to usually pertains to single-particle analysis, which requires orientation determination and averaging of hundreds of thousands of particles. This method assumes that the particles are homogeneous. However, the mRNA within LNPs does not exhibit such homogeneity, as each mRNA molecule may have a different structure. Therefore, it is not possible to perform particle alignment and averaging for these molecules.