|
1. Protein Homology Modeling
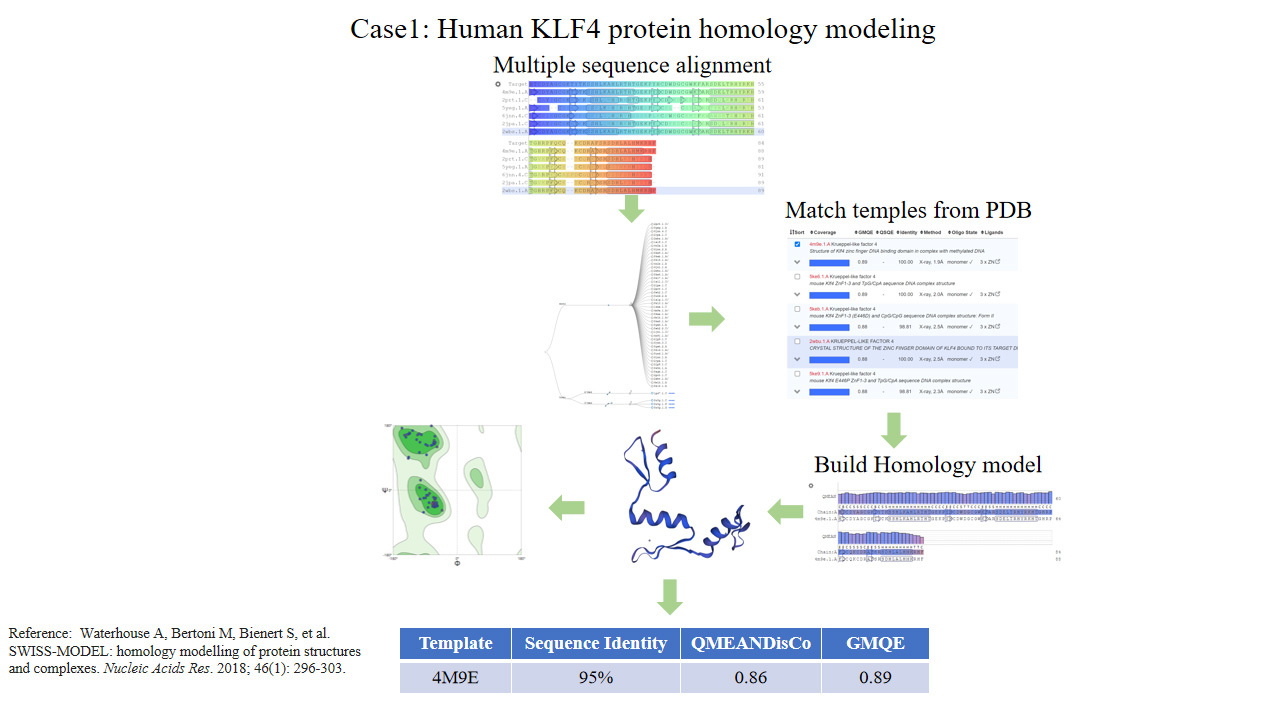
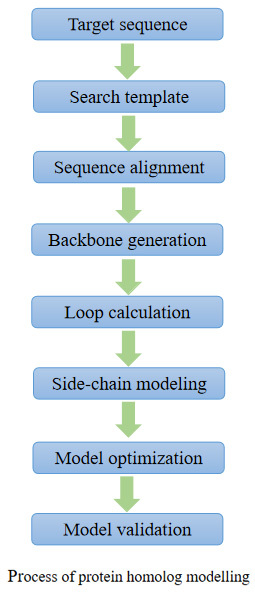
Homologous modeling could convert amino acid sequences to three-dimensional structures based on template proteins. It is currently the most effective method for predicting the three-dimensional protein structure. For unpublished protein structures, homology modeling is an important and effective structure building method. Based on the conservative theory of the three-dimensional structure of homologous proteins, providing protein sequence identity is greater than 30%, proteins with unknown structures can use one or more related structures as templates to construct their three-dimensional structures. The constructed protein model can be used for subsequent molecular mechanism studies.
2. Protein De-novo design
De novo design, also known as the "reverse protein folding problem", is an attractive method for constructing proteins with predetermined structures and functions. The basis of de-novo design is to identify the primary sequence that can be correctly folded into desired structural topology. After an in-depth understanding of protein structure and related functions, backbone construction is the first but an essential step, in which minor elements such as α-helices, β-chains and loops are assembled with the help of machine learning and algorithms. De-novo design helps to generate the appropriate sequence and optimize the desired conformation. Biortus team can provide de-novo design related services for protein engineering, including protein structure prediction, protein folding and interaction, stability prediction etc.
|
 |